Medea Vasp Download
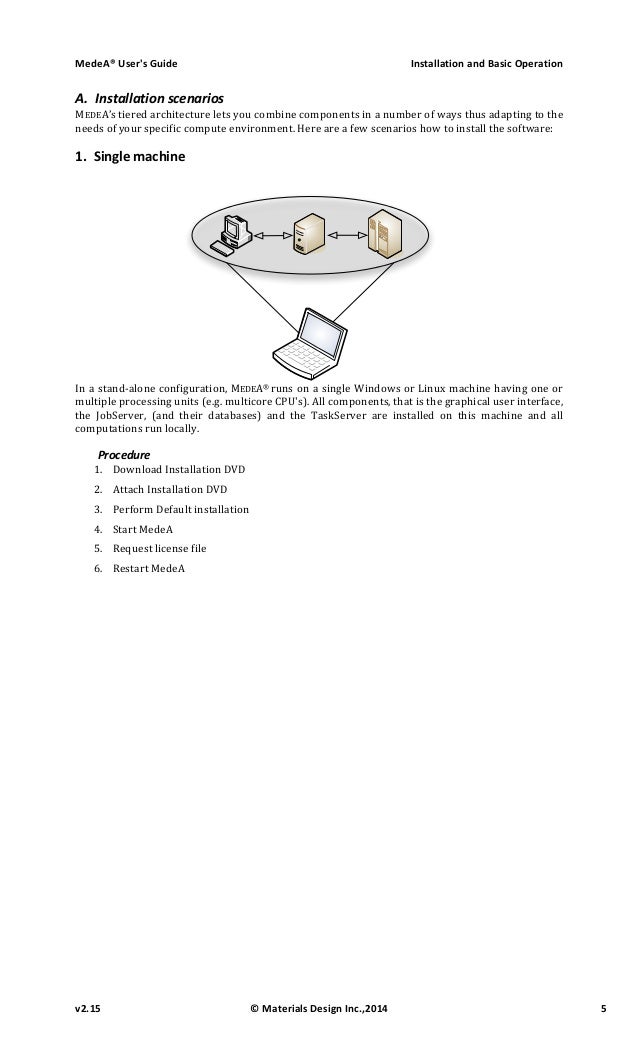
. Lots of functionality, specific VASP options e.g. Dinosaur king arcade game emulator. Download fcc Ag.cif from Crystallography Open Database and test: cif2cell 9008459.cif -p vasp -vasp-format=5 -supercell=2,2,2. Possessing a comprehensive array of advanced features, including hybrid functionals, the ability to incorporate dispersion interactions, and comprehensive and validated self-consistent PAW potentials, MedeA VASP 5 provides access to state of the art first-principles. The vacancy is in the +2. © 2020 by Materials Design, Inc. Privacy Policy Materials Design® and MedeA® are registered trademarks of Materials Design, Inc. MedeA ® VASP 5.2 VASP 5.2, first released in mid 2009, represents a breakthrough in the calculation of electronic and optical properties for semiconductors and insulators of industrial importance. This is based on an efficient implementation of hybrid functionals and the GW methods.
Requests for technical support from the VASP group should be posted in the VASP-forum.
- 2Input
- 3Calculation
Task
In this example the nucleophile substitution of a Cl- by another Cl- in CH3Cl is simulated using bluemoon sampling.
Medea Vasp Download Free
Input

POSCAR
In the blue moon sampling method several POSCAR files are used for different values of the collective variable.
Vasp Download Linux
KPOINTS
- For isolated atoms and molecules interactions between periodic images are negligible (in sufficiently large cells) hence no Brillouin zone sampling is necessary.
INCAR
- The setting LBLUEOUT=.TRUE. tells VASP to write out the information needed for the computation of free energies.

Calculation
In the blue moon method, the free energy difference is computed by integration of the free energy gradients computed for several points differing in the value of the collective variable distributed between known inital and final states. The gradients for each point are computed within a constrained molecular dynamics simulation (note the value of STATUS=0 for the collective variable defined in the ICONST file).
Running the calculation
The mass for hydrogen in this example is set 3.016 a.u. corresponding to the tritium isotope. This way larger timesteps can be chosen for the MD (note that the free energy is independent of the masses of atoms). The simulation for each of the points along the reaction coordinate is performed in a separate directory called 1, 2, .., 7. These are created automatically by the run script. For practical reasons, we split our (presumably long) blue moon calculation into shorter runs of length of 1000 fs (NSW=1000; POTIM=1). This is done automatically in the script run which looks as follows:
Free-energy profile
The free energy gradient is obtained as a ratio of two averages (see Constrained molecular dynamics). This is done by the script fgradBM.sh, which writes the free energy gradient vs. the collective variable to the file grad.dat:
To execute that script type: Sims 3 get high mod.
For our purposes, a simple trapezoidal rule can be used for the integration of gradients. For accurate calculations, more sohpisticated integration schemes should be considered.The free energy vs. collective variable is obtained by forward integration using the script integrateForward.py:
To execute that script type and write to the file free_energy.dat:
Finally to plot that file type:
The free energy profile should look like the following:

Medea Vasp Download Torrent
Note that much longer simulations should be performed (typically a few tens or hundreds of ps) in order to achieve well converged averages needed in accurate calculations.